Genome Mining
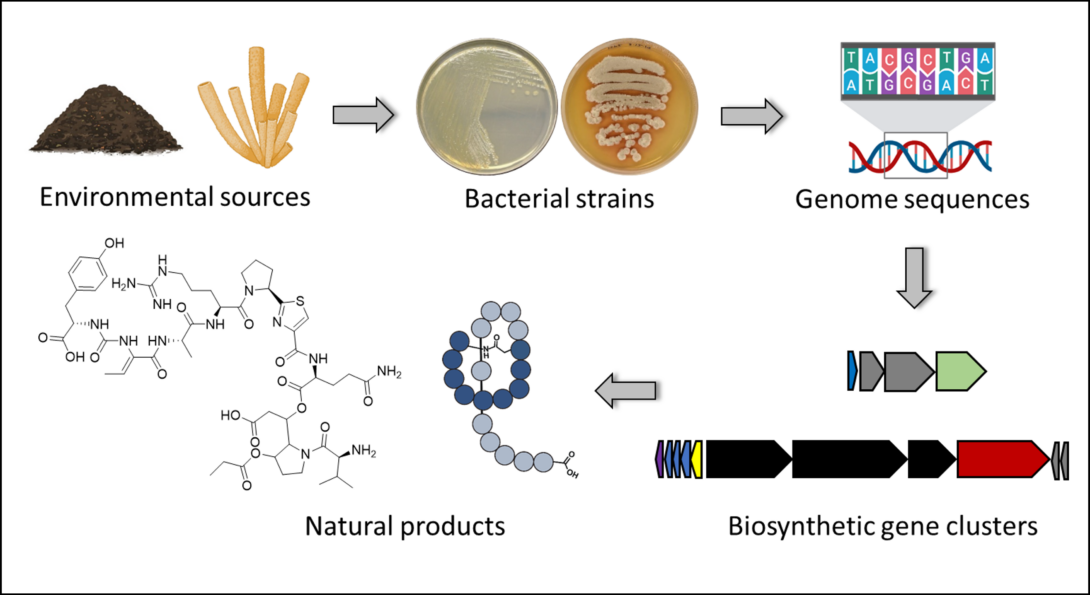
Discovering the Biosynthetic Potential of Bacteria
The ability to predict the biosynthetic potential of bacteria from their genome sequences (“from genes to molecules”) and the ability to predict which genes code for the biosynthesis of a particular natural product (“from molecules to genes”) has the potential to revolutionize drug discovery and development efforts:
1) With the increasing number of microbial genome sequences available, it has become clear that the biosynthetic potential of microorganisms is much higher than what we observe through fermentation. A typical, natural product-producing bacterium will be known to make 1-3 natural products, but ~30 may be predicted from its genome sequence. Why this discrepancy? Because most biosynthetic genes are silent or not well expressed under laboratory conditions. But knowing which genes are there (knowing what the potential is) gives you the chance to activate those genes and get the corresponding natural products.
2) Imagine you have a strain collection of 1,000s to 100,000s of strains. The ability to predict what types of natural products each strain can potentially make through genome mining, gives you the opportunity to select potentially “talented” strains to be included in the natural product discovery pipeline.
3) Identifying the genes that encode the biosynthesis of a natural product of interest gives you the opportunity to engineer the biosynthesis for yield improvement or structure diversification.
References
- Paulo BS, Recchia MJJ, Lee S, Fergusson CH, Romanowski SB, Hernandez A, Krull N, Liu DY, Cavanagh H, Bos A, Gray CA, Murphy BT, Linington RG, Eustaquio AS* (2024) Discovery of megapolipeptins by genome mining of a Burkholderiales bacteria collection. Chem Sci 15(40):16567–81. doi: 10.1039/d4sc03594a.
- Romanowski SB, Lee S, Kunakom S, Paulo BS, Recchia MJJ, Liu DY, Cavanagh H, Linington RG, Eustáquio AS* (2023) Identification of the lipodepsipeptide selethramide encoded in a giant nonribosomal peptide synthetase from a Burkholderia bacterium. Proc Natl Acad Sci U S A 120(42):e2304668120. doi: 10.1073/pnas.2304668120.
- Ióca LP, Dai Y, Kunakom S, Diaz-Espinosa J, Krunic A, Crnkovic CM, Orjala J, Sanchez LM, Ferreira AG, Berlinck RGS, Eustáquio AS* (2021) A Family of Nonribosomal Peptides Modulate Collective Behavior in Pseudovibrio Bacteria Isolated from Marine Sponges. Angew Chem Int Ed Engl 60(29):15891-15898. doi: 10.1002/anie.202017320.
- Braesel J, Arnould B, Lee J, Murphy BT, Eustáquio AS* (2019) Diazaquinomycin biosynthetic gene clusters from marine and freshwater actinomycetes. J Nat Prod 82: 937-946.
- Braesel J, Crnkovic CM, Kunstman KJ, Green SJ, Maienschein-Cline M, Orjala J, Murphy BT, Eustáquio AS* (2018) Complete genome of Micromonospora sp. strain B006 reveals biosynthetic potential of a Lake Michigan actinomycete. J Nat Prod 81: 2057-2068.