Research Interests
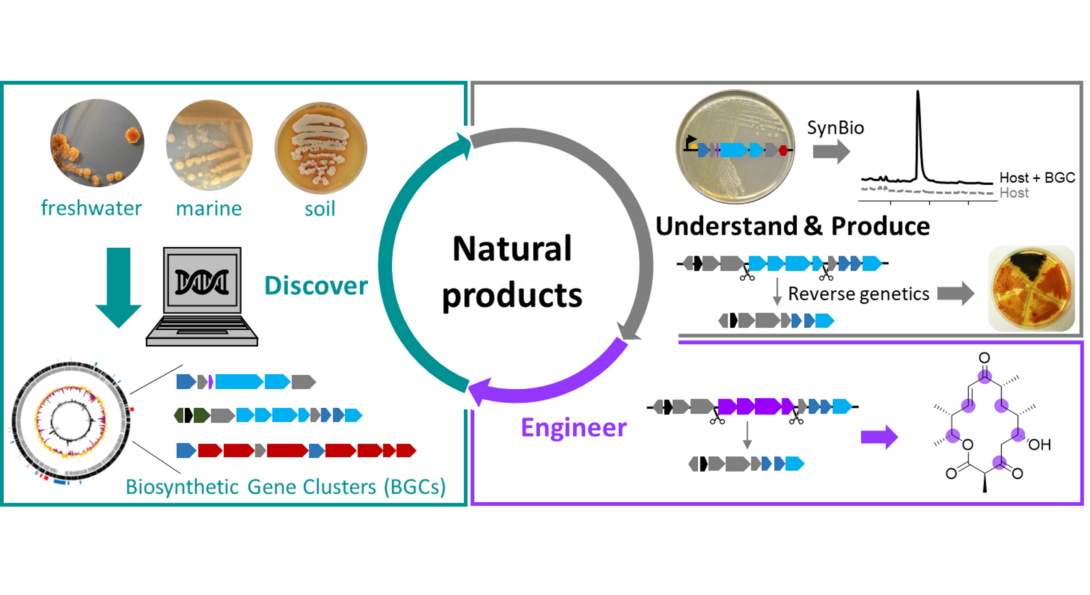
Introduction
We are interested in studying natural products (specialized metabolites) produced by bacteria. Natural products play a crucial role in human health. For example, more than half of the antibiotics used today originate from environmental bacteria. However, challenges include rediscovery, limited access from natural sources, and difficulties in introducing structural modifications necessary to convert a natural product into a pharmaceutical.
Our laboratory tackles these challenges by:
1) Applying genome mining,
2) Developing synthetic biology tools,
3) Establishing reverse genetics for non-model bacteria
4) Via biosynthetic engineering.
References
- Kunakom & Eustáquio (2019) Natural products and synthetic biology – where we are and where we need to go. mSystems 4:e00113-19.
- Adaikpoh, Fernandez & Eustáquio (2022) Biotechnology approaches for natural product discovery, engineering and production based on Burkholderia. Curr Opin Biotechnol 77:102782.